[](https://github.com/cjneely10/affinityprop/actions/workflows/rust.yml)
[](https://crates.io/crates/affinityprop)
[](https://docs.rs/affinityprop/0.2.0/affinityprop/)
[](https://www.gnu.org/licenses/gpl-3.0.html)
[](https://doc.rust-lang.org/rustc/what-is-rustc.html)

# affinityprop
The `affinityprop` crate provides an optimized implementation of the Affinity Propagation
clustering algorithm, which identifies cluster of data without *a priori* knowledge about the
number of clusters in the data. The original algorithm was developed by
Brendan Frey and Delbery Dueck
# About
Affinity Propagation identifies a subset of representative examples from a dataset, known as
**exemplars**.
Briefly, the algorithm accepts as input a matrix describing **pairwise similarity** for all data
values. This information is used to calculate pairwise **responsibility** and **availability**.
Responsibility *`r(i,j)`* describes how well-suited point *`j`* is to act as an exemplar for
point *`i`* when compared to other potential exemplars. Availability *`a(i,j)`* describes how
appropriate it is for point *i* to accept point *j* as its exemplar when compared to
other exemplars.
Users provide a number of **convergence iterations** to repeat the calculations, after which the
potential exemplars are extracted from the dataset. Then, the algorithm continues to repeat
until the exemplar values stop changing, or until the **maximum iterations** are met.
# Why this crate?
The nature of Affinity Propagation demands an *O(n2)* runtime. An existing
sklearn
version is implemented using the Python library
numpy
which incorporates vectorized row operations. Coupled with **SIMD** instructions, this results
in decreased time to finish.
However, in applications with large input values, the *O(n2)* runtime is still
prohibitive. This crate implements Affinity Propagation using the
rayon
crate, which allows for a drastic decrease in overall runtime - as much as 30-60% when compiled
in release mode!
# Dependencies
cargo
with `rustc >=1.58`
# Installation
## In Rust code
```toml
[dependencies]
affinityprop = "0.2.0"
ndarray = "0.15.4"
```
## As a command-line tool
```shell
cargo install affinityprop
```
# Usage
## From Rust code
The `affinityprop` crate expects a type that defines how to calculate pairwise `Similarity`
for all data points. This crate provides the `NegEuclidean`, `NegCosine`, and
`LogEuclidean` structs, which are defined as `-1 * sum((a - b)**2)`, `-1 * (a . b)/(|a|*|b|)`,
and `sum(log((a - b)**2))`, respectively.
Users who wish to calculate similarity differently are advised that **Affinity Propagation
expects *s(i,j)* > *s(i, k)* iff *i* is more similar to *j* than it is to *k***.
```rust
use ndarray::{arr1, arr2, Array2};
use affinityprop::{AffinityPropagation, NegCosine, Preference};
let x: Array2 = arr2(&[[0., 1., 0.], [2., 3., 2.], [3., 2., 3.]]);
// Cluster using negative cosine similarity with a pre-defined preference
let ap = AffinityPropagation::default();
let (converged, results) = ap.predict(&x, NegCosine::default(), Preference::Value(-10.));
assert!(converged && results.len() == 1 && results.contains_key(&0));
// Cluster with list of preference values
let pref = arr1(&[0., -1., 0.]);
let (converged, results) = ap.predict(&x, NegCosine::default(), Preference::List(&pref));
assert!(converged);
assert!(results.len() == 2 && results.contains_key(&0) && results.contains_key(&2));
// Use damping=0.5, threads=2, convergence_iter=10, max_iterations=100,
// median similarity as preference
let ap = AffinityPropagation::new(0.5, 2, 10, 100);
let (converged, results) = ap.predict(&x, NegCosine::default(), Preference::Median);
assert!(converged);
assert!(results.len() == 2 && results.contains_key(&0) && results.contains_key(&2));
// Predict with pre-calculated similarity
let s: Array2 = arr2(&[[0., -3., -12.], [-3., 0., -3.], [-12., -3., 0.]]);
let ap = AffinityPropagation::default();
let (converged, results) = ap.predict_precalculated(s, Preference::Value(-10.));
assert!(converged && results.len() == 1 && results.contains_key(&1));
```
## From the Command Line
`affinityprop` can be run from the command-line and used to analyze a file of data:
```text
ID1 val1 val2
ID2 val3 val4
ID3 val5 val6
```
where ID*n* is any string identifier and val*n* are floating-point (decimal) values.
The file delimiter is provided from the command line with the `-l` flag.
Similarity will be calculated based on the option set by the `-s` flag.
For files without row ids:
```text
val1 val2
val3 val4
val5 val6
```
provide the `-n` flag from the command line. IDs will automatically be assigned by zero-based
index.
Users may instead provide a pre-calculated similarity matrix by passing the `-s 3` flag from
the command line and by structuring their input file as:
```text
ID1 sim11 sim12 sim13
ID2 sim21 sim22 sim23
ID3 sim31 sim32 sim33
```
where row*i*, col*j* is the pairwise similarity between inputs *i* and *j*.
Or, for files without row labels, users may pass `-n -s 3`:
```text
sim11 sim12 sim13
sim21 sim22 sim23
sim31 sim32 sim33
```
IDs will automatically be assigned by zero-based index.
### Help Menu
```text
affinityprop 0.2.0
Chris N.
Vectorized and Parallelized Affinity Propagation
USAGE:
affinityprop [OPTIONS] --input
FLAGS:
-n, --no_labels Input file does not contain IDS as the first column
-h, --help Prints help information
-V, --version Prints version information
OPTIONS:
-c, --convergence_iter Convergence iterations, default=10
-d, --damping Damping value in range (0, 1), default=0.9
-l, --delimiter File delimiter, default '\t'
-i, --input Path to input file
-m, --max_iter Maximum iterations, default=100
-r, --precision Set f32 or f64 precision, default=f32
-p, --preference Preference to be own exemplar, default=median pairwise similarity
-s, --similarity Set similarity calculation method
(0=NegEuclidean,1=NegCosine,2=LogEuclidean,3=precalculated), default=0
-t, --threads Number of worker threads, default=4
```
### Results
Results are printed to stdout in the format:
```text
Converged=true/false nClusters=NC nSamples=NS
>Cluster=n size=N exemplar=i
[comma-separated cluster member IDs/indices]
>Cluster=n size=N exemplar=i
[comma-separated cluster member IDs/indices]
...
```
# Runtime and Resource Notes
Affinity Propagation is *O(n2)* in both runtime and memory.
This crate seeks to address the former, not the latter.
An estimated memory usage can be calculated given:
```text
memory(GB) = p * 4 * N^2 / 2^30
```
For `N` inputs. `p = 4` for 32-bit floating-point precision and `p = 8` for 64-bit.
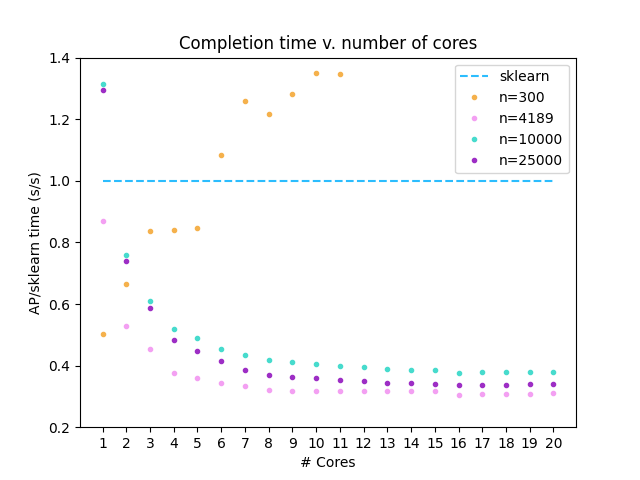
## Comparison to sklearn implementation
This implementation was tested on 50-D isotropic Gaussian blobs generated by the sklearn
[make_blobs](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.make_blobs.html)
function (n=300,10000,25000). A [dataset of biological relevance](https://github.com/edgraham/BinSanity/blob/master/example/Infant_gut_assembly.cov.x100.lognorm)
was also selected (n=4189). ARI/F1 scores >= 0.99 were obtained for the Gaussian data when compared to the labels
provided by `make_blobs`. ARI/F1 = 0.98 was obtained for the biological dataset when compared to `sklearn`-derived
labels.
This `affinityprop` implementation was compared against the [Affinity Propagation](https://scikit-learn.org/stable/modules/generated/sklearn.cluster.AffinityPropagation.html#sklearn.cluster.AffinityPropagation.fit_predict)
implementation contained within scikit-learn-1.0.2, and run using numpy-1.22.2 with Python 3.10.0.
This analysis was completed using a Ryzen 9 3950X processor.
In all analyses, damping=0.95, convergence_iter=400, and max_iter=4000.
Preference=-1000.0 for Gaussian data and -10.0 for biological data.