[](https://github.com/wh-xu/Hyper-Gen/blob/main/LICENSE)
## HyperGen: Compact and Efficient Genome Sketching using Hyperdimensional Vectors
_HyperGen_ is a Rust library used to sketch genomic files and realize fast Average Nucleotide Identity (ANI) approximation. _HyperGen_ leverages two advanced algorithms: 1. FracMinHash and 2. hyperdimensional computing (HDC) with random indexing as shown in the following figure:
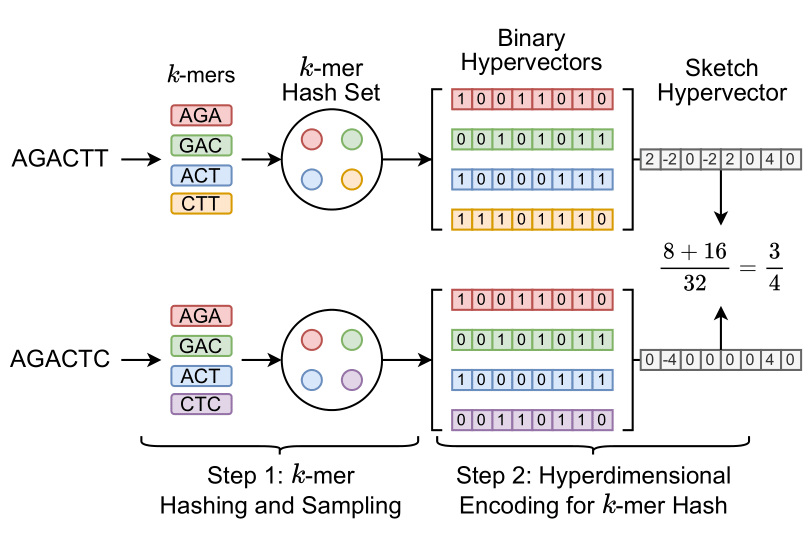
_HyperGen_ first samples the kmer set using FracMinHash. Then the kmer hashes are encoded into hyperdimensional vectors (HVs) using HDC encoding to obtain better tradeoff of ANI estimation quality, sketch size, and computation speed. The sketch size generated by _HyperGen_ is 1.8 to 2.7× smaller than _Mash_ and _Dashing 2_. ANI estimation in _HyperGen_ can be realized using highly vectorized vector multiplication. _HyperGen_'s database search speed for large-scale datasets is up to 4.3x faster than _Dashing 2_.
## Quickstart
### Installation
#### Basic Installation
_HyperGen_ requires [`Rust`](https://www.rust-lang.org/tools/install) language and [`Cargo`](https://doc.rust-lang.org/cargo/) to be installed.
We recommend installing _HyperGen_ using the following command:
```sh
git clone https://github.com/wh-xu/Hyper-Gen.git
cd Hyper-Gen
# Without GPU acceleration for sketching
cargo install --path .
```
#### Install with GPU Support
_HyperGen_ supports GPU acceleration. Using GPU mode will require the installation of NVIDIA GPU driver. Use `nvidia-smi` or `nvcc -V` to check if the driver is installed. Then run the following command to install with GPU support:
```sh
# With GPU acceleration for sketching
cargo install --features cuda-sketch --path .
```
Currently only Nvidia GPUs are supported. We tested the compatibility on both desktop `RTX4090` and laptop `RTX4060` with CUDA Version `12.x`.
### Usage
Current version supports following functions:
#### 1. Genome sketching for .fa/.fna/.fasta files
```sh
Example:
hyper-gen sketch -p ./data -o ./fna.sketch
Positional arguments:
-p, --path Input folder path to sketch
-o, --out Output path
-t, --thread Threads used for computation [default: 16]
-C, --canonical If use canonical kmer [default: true]
-k, --ksize k-mer size for sketching [default: 21]
-s, --scaled Scaled factor for FracMinHash [default: 1500]
-d, --hv_d Dimension for hypervector [default: 4096]
-D, --device Device to run [default: cpu] [possible values: cpu, gpu]
```
#### 2. ANI estimation and database search
```sh
Example:
hyper-gen dist -r fna1.sketch -q fna2.sketch -o output.ani
Positional arguments:
-r, --path_r Path to ref sketch file
-q, --path_q Path to query sketch file
-o, --out Output path
-t, --thread Threads used for computation [default: 16]
-a, --ani_th ANI threshold [default: 85.0]
```
#### 3. Faster sketching on GPU
_HyperGen_ supports offloading the kmer hashing and sampling steps to GPU to speed up the sketching process. Use the following command to run on GPU device:
```sh
hyper-gen sketch -D gpu -p ./data -o ./fna.sketch
```
## Differences between _Mash_ and _HyperGen_
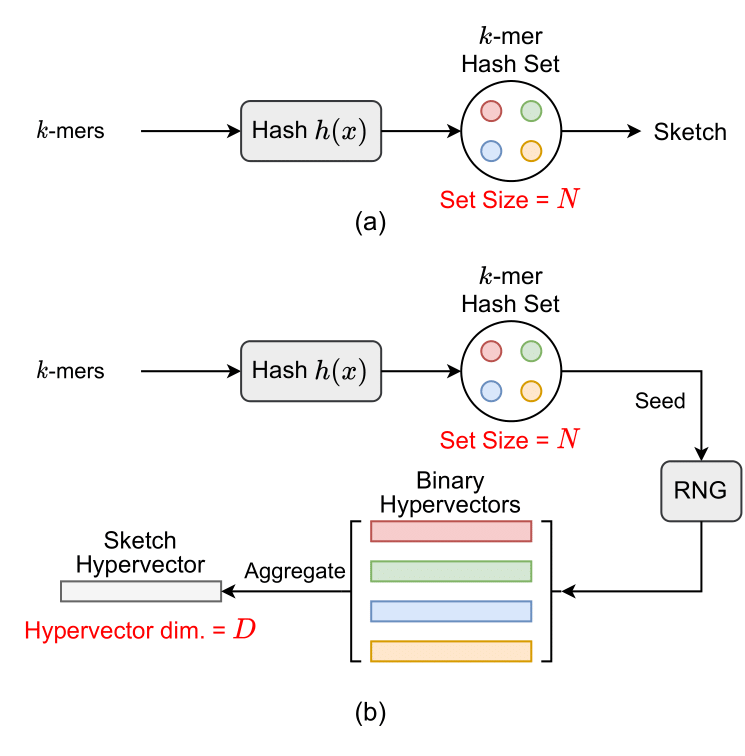
- (a) _Mash_ uses MinHash to sample kmer hash set and stores discrete hash values as the genome sketch.
- (b) _HyperGen_ uses FracMinHash to sample kmer hash set and encodes discrete hash values into continuous _sketch hypervector_.
## Publication
1. Weihong Xu, Po-kai Hsu, Niema Moshiri, Shimeng Yu, and Tajana Rosing. "[HyperGen: Compact and Efficient Genome Sketching using Hyperdimensional Vectors](https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btae452/7714688)." _Bioinformatics_, 2024.
## Contact
For more information, post an issue or send an email to .


