neighborlist-rs
| Crates.io | neighborlist-rs |
| lib.rs | neighborlist-rs |
| version | 0.1.9 |
| created_at | 2026-01-11 02:45:36.795834+00 |
| updated_at | 2026-01-11 04:09:14.712409+00 |
| description | High-performance neighborlist construction for atomistic systems |
| homepage | |
| repository | |
| max_upload_size | |
| id | 2035123 |
| size | 6,659,356 |
documentation
README
neighborlist-rs
High-performance neighborlist construction for atomistic systems in Rust with Python bindings.
Features
- High-performance Cell Lists: O(N) scaling for large systems.
- Batched Processing: Parallelized neighbor list construction across multiple systems (ideal for GNN training).
- Multi-Cutoff pass: Generate multiple neighbor lists (e.g., short-range GNN, long-range dispersion) in a single optimized pass.
- SIMD Acceleration: Explicit
wideSIMD kernels for small systems and brute-force fallbacks. - Auto-Box Inference: Automatic bounding box calculation for isolated molecules.
- Robust PBC support: Minimum image convention for orthorhombic and triclinic cells.
- ASE Integration: Direct support for
ase.Atomsobjects. - Zero-copy NumPy integration: Efficient data exchange via PyO3 and
rust-numpy.
Installation
From PyPI
pip install neighborlist-rs
From Source
pip install .
Usage (Python)
Refer to PYTHON_API.md for full documentation and advanced usage examples.
Quick Start (with ASE)
import neighborlist_rs
from ase.build import molecule
atoms = molecule("C2H5OH")
# Single pass search directly from ASE Atoms
result = neighborlist_rs.build_from_ase(atoms, cutoff=5.0)
edge_index = result["edge_index"] # (2, M) array of atom pairs
shifts = result["shift"] # (M, 3) periodic shift vectors
Manual Search
import neighborlist_rs
import numpy as np
positions = np.array([[0, 0, 0], [1, 1, 1]], dtype=float)
# None for cell means isolated system (non-PBC)
result = neighborlist_rs.build_neighborlists(None, positions, cutoff=5.0)
Usage (Rust)
Refer to RUST_API.md for detailed Rust documentation.
Single System Search
use neighborlist_rs::build_neighborlists;
let positions = [[0.0, 0.0, 0.0], [1.0, 0.0, 0.0]];
let cutoff = 1.5;
let result = build_neighborlists(&positions, cutoff, None, true).unwrap();
Performance
neighborlist-rs is designed for high-performance atomistic simulations, outperforming established baselines across a wide range of system sizes and conditions.
Comprehensive Benchmarks
(Measured on CPU: ARM64 8-core / NVIDIA GB10 proxy. Results in wall-clock time ms.)
1. Single System Scaling
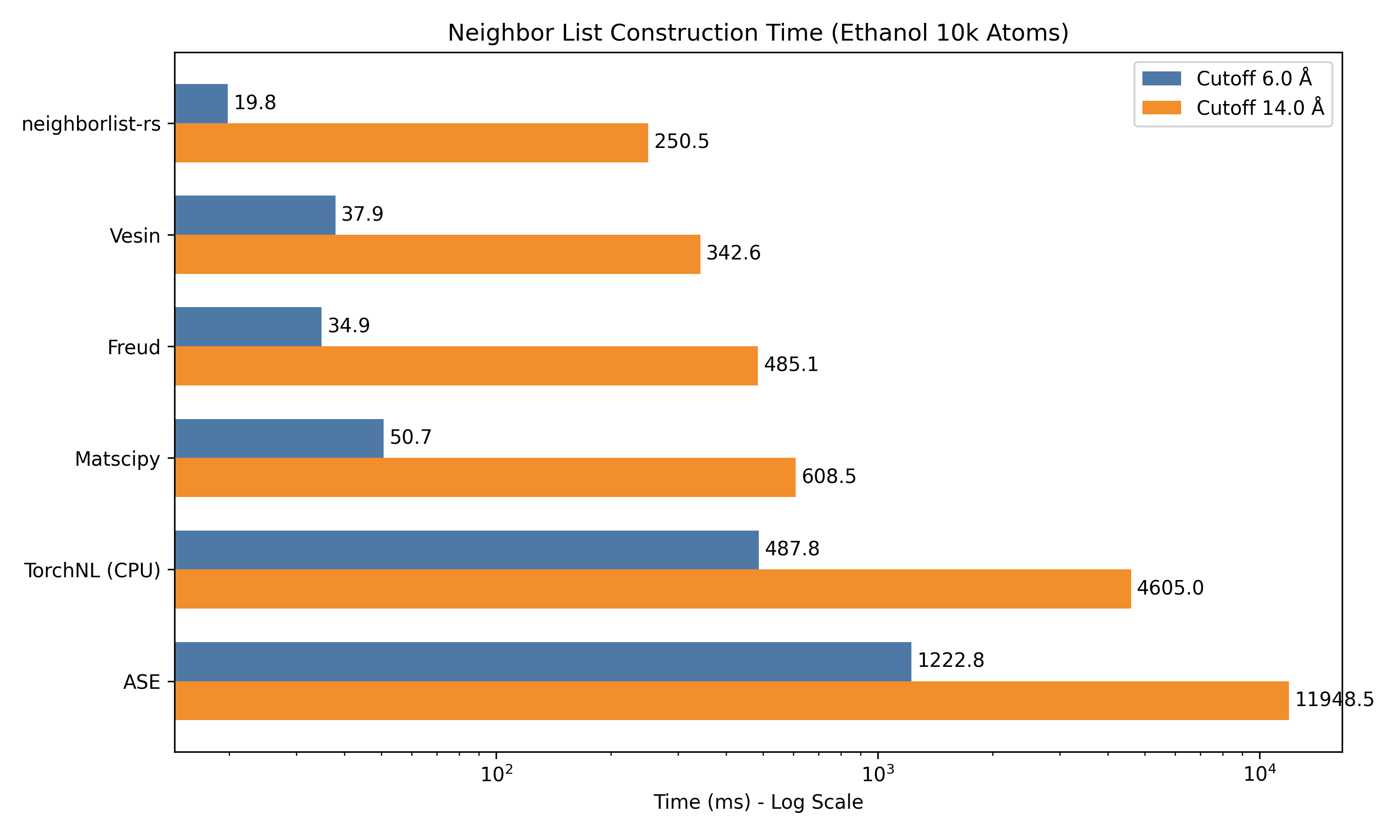
The table below shows the average wall-clock time (in milliseconds) to construct the neighbor list. Lower is better.
| System | Cutoff | ASE | Matscipy | Freud | Vesin | neighborlist-rs |
|---|---|---|---|---|---|---|
| Isolated 1k | 6.0 Å | 101.5 | 3.3 | 4.1 | 1.7 | 3.0 |
| 14.0 Å | 2688.1 | 10.2 | 12.2 | 5.8 | 2.8 | |
| Ethanol 1k | 6.0 Å | 149.9 | 5.9 | 2.7 | 3.0 | 5.3 |
| (PBC) | 14.0 Å | 6677.0 | 130.8 | N/A* | 64.0 | 37.6 |
| Ethanol 10k | 6.0 Å | 1222.8 | 50.7 | 34.9 | 37.9 | 19.8 |
| (PBC) | 14.0 Å | 11948.5 | 608.5 | 485.1 | 342.6 | 250.5 |
*Freud requires box dimensions > 2x cutoff.
Detailed Raw Timing Summary
======================================== Single System Benchmarks ========================================
>>> CUTOFF: 6.0 Angstroms <<<
--- Isolated 1k (N=1000, rc=6.0) ---
Library | Device | Time (ms) | Std
-------------------------------------------------------
ASE | CPU | 101.48 | 0.76
Matscipy | CPU | 3.28 | 0.01
Freud | CPU | 4.13 | 0.85
Vesin | CPU | 1.72 | 0.02
RS (1 thread) | CPU | 3.16 | 0.47
RS (Parallel) | CPU | 2.97 | 0.26
TorchCluster | CPU | 2.49 | 0.11
TorchCluster | GPU | 18.51 | 1.05
TorchNL | CPU | 16.34 | 3.01
TorchNL | GPU | 6.52 | 0.80
--- Ethanol 1k (PBC) (N=1008, rc=6.0) ---
Library | Device | Time (ms) | Std
-------------------------------------------------------
ASE | CPU | 149.92 | 15.19
Matscipy | CPU | 5.91 | 0.01
Freud | CPU | 2.71 | 0.59
Vesin | CPU | 2.99 | 0.02
RS (1 thread) | CPU | 5.44 | 0.11
RS (Parallel) | CPU | 5.35 | 0.01
TorchCluster | --- | N/A (No PBC support) |
TorchNL | CPU | 31.72 | 1.27
TorchNL | GPU | 18.85 | 4.69
--- Ethanol 10k (PBC) (N=10008, rc=6.0) ---
Library | Device | Time (ms) | Std
-------------------------------------------------------
ASE | CPU | 1222.77 | 2.29
Matscipy | CPU | 50.70 | 0.21
Freud | CPU | 34.93 | 2.36
Vesin | CPU | 37.92 | 0.56
RS (1 thread) | CPU | 20.61 | 0.68
RS (Parallel) | CPU | 19.82 | 0.10
TorchCluster | --- | N/A (No PBC support) |
TorchNL | CPU | 487.77 | 8.08
TorchNL | GPU | 134.06 | 27.62
>>> CUTOFF: 14.0 Angstroms <<<
--- Isolated 1k (N=1000, rc=14.0) ---
Library | Device | Time (ms) | Std
-------------------------------------------------------
ASE | CPU | 2688.05 | 5.74
Matscipy | CPU | 10.16 | 0.03
Freud | CPU | 12.20 | 2.00
Vesin | CPU | 5.82 | 0.06
RS (1 thread) | CPU | 2.89 | 0.07
RS (Parallel) | CPU | 2.82 | 0.06
TorchCluster | CPU | 9.14 | 0.51
TorchCluster | GPU | 18.36 | 0.82
TorchNL | CPU | 81.01 | 4.50
TorchNL | GPU | 24.08 | 1.98
--- Ethanol 1k (PBC) (N=1008, rc=14.0) ---
Library | Device | Time (ms) | Std
-------------------------------------------------------
ASE | CPU | 6676.95 | 4.42
Matscipy | CPU | 130.84 | 0.63
Vesin | CPU | 63.95 | 0.47
RS (1 thread) | CPU | 37.62 | 0.22
RS (Parallel) | CPU | 37.88 | 0.15
TorchCluster | --- | N/A (No PBC support) |
TorchNL | CPU | 522.08 | 8.28
TorchNL | GPU | 90.39 | 2.55
--- Ethanol 10k (PBC) (N=10008, rc=14.0) ---
Library | Device | Time (ms) | Std
-------------------------------------------------------
ASE | CPU | 11948.54 | 273.39
Matscipy | CPU | 608.45 | 2.64
Freud | CPU | 485.05 | 3.00
Vesin | CPU | 342.63 | 1.84
RS (1 thread) | CPU | 250.45 | 5.98
RS (Parallel) | CPU | 264.32 | 3.57
TorchCluster | --- | N/A (No PBC support) |
TorchNL | CPU | 4605.02 | 71.68
TorchNL | GPU | 790.87 | 9.78
2. Multi-Cutoff Pass [6.0, 14.0] Å
Benchmark for generating two neighbor lists simultaneously (e.g., GNN + Dispersion). (System: Ethanol 10k atoms)
| Library | Strategy | Time (ms) |
|---|---|---|
| neighborlist-rs | Single Pass (Native) | 313 |
| Vesin | Filtered (14.0 -> 6.0) | 414 |
| Freud | Filtered (14.0 -> 6.0) | 491 |
| Matscipy | Filtered (14.0 -> 6.0) | 667 |
| ASE | Filtered (14.0 -> 6.0) | 11680 |
3. Batch Throughput (GNN Workloads)
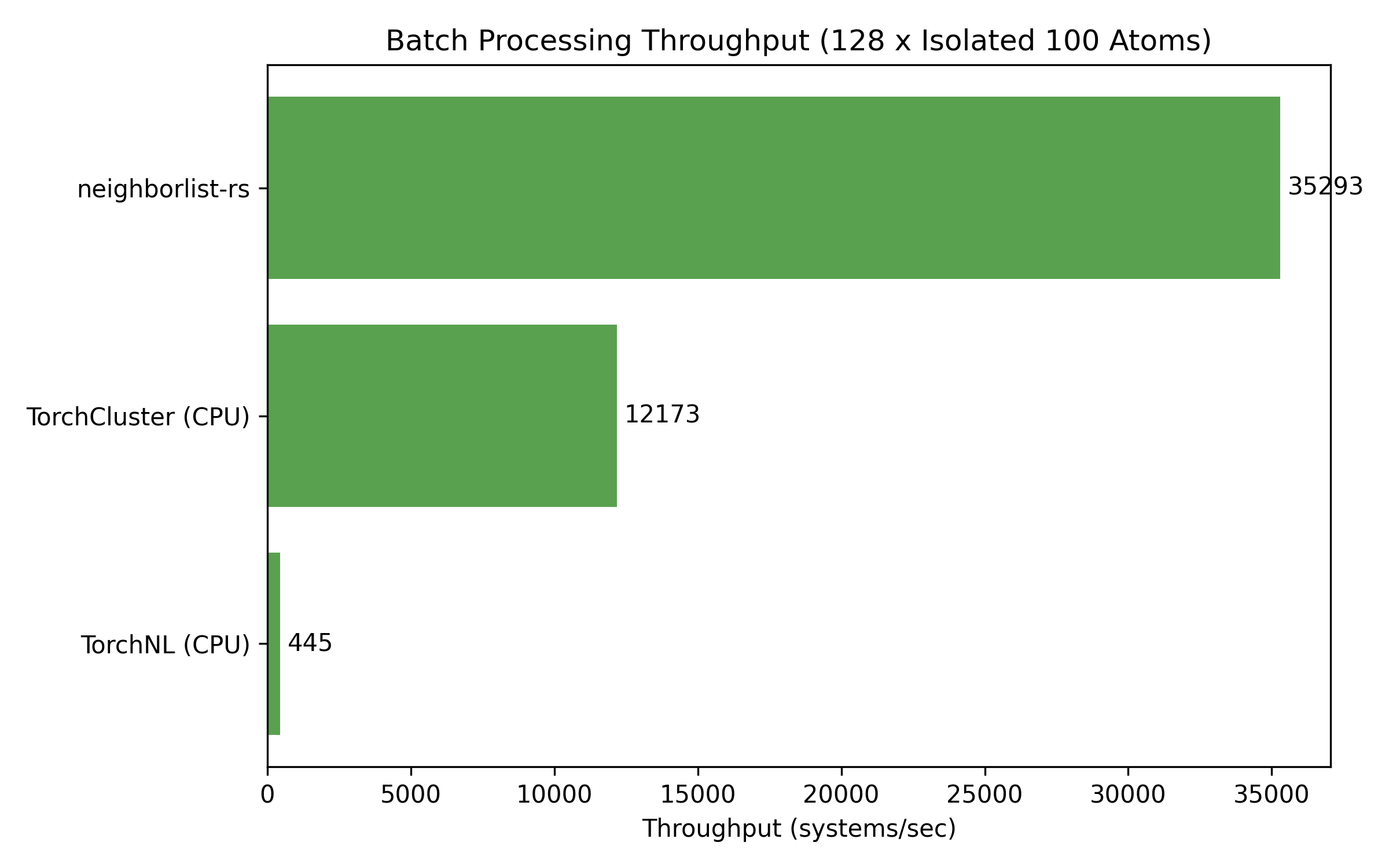
Throughput in systems per second for batches of isolated 100-atom molecules.
| Batch Size | neighborlist-rs | TorchCluster | TorchNL |
|---|---|---|---|
| 1 | 34,477 | 11,756 | 525 |
| 32 | 38,198 | 11,047 | 481 |
| 128 | 35,293 | 12,173 | 445 |
Note: TorchCluster does not support Periodic Boundary Conditions (PBC).
Key Optimizations
- Spatial Sorting: Uses Z-order (Morton) indexing to improve cache locality.
- Dynamic Configuration: Runtime-tunable thresholds for switching between SIMD Brute-Force and Cell Lists.
- Two-Pass Search: Minimizes heap allocations by pre-calculating neighbor counts.
- Cross-Platform SIMD: Optimized for both x86_64 (AVX2/FMA) and aarch64 (NEON).
Verification
Run Rust tests:
cargo test
Run Python tests:
pytest tests/
License
Licensed under either of:
- Apache License, Version 2.0 (LICENSE-APACHE or http://www.apache.org/licenses/LICENSE-2.0)
- MIT license (LICENSE-MIT or http://opensource.org/licenses/MIT) at your option.

