stree_cmd
| Crates.io | stree_cmd |
| lib.rs | stree_cmd |
| version | 0.2.1 |
| created_at | 2015-06-19 23:56:31.2643+00 |
| updated_at | 2015-12-11 23:56:18.321051+00 |
| description | Command line tool for creating and visualizing suffix trees. |
| homepage | https://github.com/BurntSushi/suffix |
| repository | https://github.com/BurntSushi/suffix |
| max_upload_size | |
| id | 2425 |
| size | 3,917 |
documentation
README
Fast linear time & space suffix arrays for Rust. Supports Unicode!
Dual-licensed under MIT or the UNLICENSE.
Documentation
The API is mostly documented with examples: http://burntsushi.net/rustdoc/suffix/.
If you just want the details on how construction algorithm used, see the
documentation for the
SuffixTable
type. This is where you'll find info on exactly how much overhead is required.
Installation
This crate works with Cargo and is on
crates.io. The package is regularly updated.
Add it to your Cargo.toml like so:
[dependencies]
suffix = "0.3"
Examples
Usage is simple. Just create a suffix array and search:
extern crate suffix;
use suffix::SuffixTable;
fn main() {
let st = SuffixTable::new("the quick brown fox was quick.");
assert_eq!(st.positions("quick"), vec![4, 24]);
}
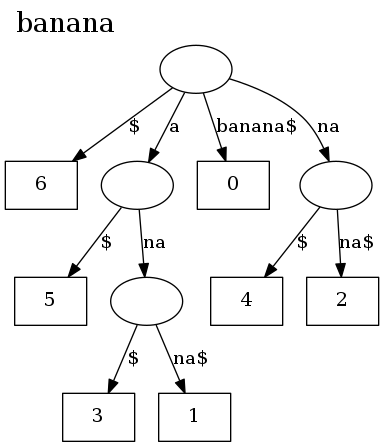
There is also a command line program, stree, that can be used to visualize
suffix trees:
git clone git://github.com/BurntSushi/suffix
cd suffix/stree_cmd
cargo build --release
./target/release/stree "banana" | dot -Tpng | xv -
And here's what it looks like:
Status of implementation
The big thing missing at the moment is a generalized suffix array. I started out with the intention to build them into the construction algorithm, but this has proved more difficult than I thought.
A kind-of-sort-of compromise is to append your distinct texts together, and
separate them with a character that doesn't appear in your document. (This is
technically incorrect, but maybe your documents don't contain any NUL
characters.) During construction of this one giant string, you should record
the offsets of where each document starts and stops. Then build a SuffixTable
with your giant string. After searching with the SuffixTable, you can find
the original document by doing a binary search on your list of documents.
I'm currently experimenting with different techniques to do this.
Benchmarks
Here are some very rough benchmarks that compare suffix table searching with searching in the using standard library functions. Note that these benchmarks explicitly do not include the construction of the suffix table. The premise of a suffix table is that you can afford to do that once---but you hope to gain much faster queries once you do.
test search_scan_exists_many ... bench: 8,601 ns/iter (+/- 85)
test search_scan_exists_one ... bench: 12 ns/iter (+/- 0)
test search_scan_not_exists ... bench: 368,788 ns/iter (+/- 811)
test search_suffix_exists_many ... bench: 414 ns/iter (+/- 15)
test search_suffix_exists_many_contains ... bench: 303 ns/iter (+/- 4)
test search_suffix_exists_one ... bench: 250 ns/iter (+/- 1)
test search_suffix_exists_one_contains ... bench: 28 ns/iter (+/- 0)
test search_suffix_not_exists ... bench: 241 ns/iter (+/- 2)
test search_suffix_not_exists_contains ... bench: 179 ns/iter (+/- 3)
The "many" benchmarks test repeated queries that match. The "one" benchmarks test a single query that matches. The "not_exists" benchmarks test a single query that does not match. Finally, the "contains" benchmark test existence rather finding all positions.
One thing you might take away from here is that you'll get a very large performance boost if many of your queries don't match. A linear scan takes a long time to fail!
And here are some completely useless benchmarks on suffix array construction.
They compare the linear time algorithm with the naive construction algorithm
(call sort on all suffixes, which is O(n^2 * logn)).
test naive_dna_medium ... bench: 33,676,690 ns/iter (+/- 86,233)
test naive_dna_small ... bench: 2,574,217 ns/iter (+/- 11,005)
test naive_small ... bench: 425 ns/iter (+/- 6)
test sais_dna_medium ... bench: 10,398,456 ns/iter (+/- 133,742)
test sais_dna_small ... bench: 977,165 ns/iter (+/- 4,125)
test sais_small ... bench: 4,038 ns/iter (+/- 39)
These benchmarks might make you say, "Whoa, the special algorithm isn't that
much faster." That's because the data just isn't big enough. And when it is
big enough, a micro benchmark is useless. Why? Because using the naive
algorithm will just burn your CPUs until the end of the time.
It would be more useful to compare this to other suffix array implementations, but I haven't had time yet. Moreover, most (all?) don't support Unicode and instead operate on bytes, which means they aren't paying the overhead of decoding UTF-8.
